Research
Plant genome sequencing and analysis
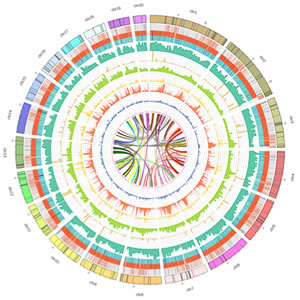 The Michael group has focused on sequencing plant genomes for over 10 years, which has resulted in over 20 plant genome papers in top journals such as Science, Nature and Cell (1–23). In addition, the Michael group published the first almost complete plant genome in Nature using single-molecule long-read Pacific Bioscience (PacBio) sequencing and BioNano Genomics optical maps (Oropetium thomaeum)(9). More recently the Michael Group has focused on Oxford Nanopore Technologies (ONT) sequencing for complete genome assemblies, direct DNA modification detection and full-length cDNA for gene prediction, and has applied this technology to generate a near complete genome version of the model plant Arabidopsis thaliana(13, 15, 20). Part of the group is concerned with developing new methods and tools for improved sequencing, while others are developing computational tools for genome analysis. While we are still building methods to generate “complete” genomes for many plant species, we are exploring computational methods to leverage all features in a genome for downstream gene discovery and molecular breeding applications for the Harnessing Plant Initiative (HPI). To this end we are developing methods for reference-free pan-genome (RFPG) analysis. Current approaches rely on a “reference” genome to anchor new genomes sequenced from the same species, which results in the loss of genomic context of many features.
The Michael group has focused on sequencing plant genomes for over 10 years, which has resulted in over 20 plant genome papers in top journals such as Science, Nature and Cell (1–23). In addition, the Michael group published the first almost complete plant genome in Nature using single-molecule long-read Pacific Bioscience (PacBio) sequencing and BioNano Genomics optical maps (Oropetium thomaeum)(9). More recently the Michael Group has focused on Oxford Nanopore Technologies (ONT) sequencing for complete genome assemblies, direct DNA modification detection and full-length cDNA for gene prediction, and has applied this technology to generate a near complete genome version of the model plant Arabidopsis thaliana(13, 15, 20). Part of the group is concerned with developing new methods and tools for improved sequencing, while others are developing computational tools for genome analysis. While we are still building methods to generate “complete” genomes for many plant species, we are exploring computational methods to leverage all features in a genome for downstream gene discovery and molecular breeding applications for the Harnessing Plant Initiative (HPI). To this end we are developing methods for reference-free pan-genome (RFPG) analysis. Current approaches rely on a “reference” genome to anchor new genomes sequenced from the same species, which results in the loss of genomic context of many features.
Time of day (TOD) expression networks
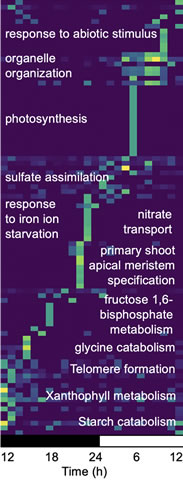 Dr. Michael’s PhD thesis was on the molecular mechanism governing the partitioning, or phasing, of biological activities to a specific time-of-day (TOD) in Arabidopsis (24–27). To extend this work, he characterized how daily light and temperature cycles interact to phase TOD expression, and found that almost all (>90%) of Arabidopsis transcripts are controlled in a TOD fashion depending on the conditions tested (28–30). Moreover, the TOD gene expression as well as the underlying transcriptional networks are conserved not only in plants but across the entire green lineage (22, 31–34). Underlying TOD expression networks is the circadian clock, which is composed of negative and positive feedback loops of a core set of genes that over evolutionary time have been codified in tight genetic linkages ensuring organisms accurately time their biology (35). Considering circadian clock genes are targets of domestication and the TOD expression networks are pleiotropic and highly conserved, the Michael group uses this as a platform to understand different aspects of plant biology. For instance, the Michael group has extended their TOD analysis suite of tools to understand water use efficiency (WUE) and Crassulacean Acid Metabolism (CAM) photosynthesis, which depends on the accurate phasing of biology to specific TOD (17, 22).
Dr. Michael’s PhD thesis was on the molecular mechanism governing the partitioning, or phasing, of biological activities to a specific time-of-day (TOD) in Arabidopsis (24–27). To extend this work, he characterized how daily light and temperature cycles interact to phase TOD expression, and found that almost all (>90%) of Arabidopsis transcripts are controlled in a TOD fashion depending on the conditions tested (28–30). Moreover, the TOD gene expression as well as the underlying transcriptional networks are conserved not only in plants but across the entire green lineage (22, 31–34). Underlying TOD expression networks is the circadian clock, which is composed of negative and positive feedback loops of a core set of genes that over evolutionary time have been codified in tight genetic linkages ensuring organisms accurately time their biology (35). Considering circadian clock genes are targets of domestication and the TOD expression networks are pleiotropic and highly conserved, the Michael group uses this as a platform to understand different aspects of plant biology. For instance, the Michael group has extended their TOD analysis suite of tools to understand water use efficiency (WUE) and Crassulacean Acid Metabolism (CAM) photosynthesis, which depends on the accurate phasing of biology to specific TOD (17, 22).
Duckweed, a minimal plant for synthetic biology
An ultimate goal of the Michael group is to develop truly synthetic plants based on the knowledge they gain from sequencing a diverse array of unique plant genomes. Therefore, the Michael group has invested in developing Duckweed as a model system since they are some of the smallest, fastest growing and simplest flowering plants known on Earth (36). Before Arabidopsis was the model plant of choice, plants in the Lemnaceae commonly known as Duckweed were important in reductionist plant biology enabling scientists to establish fundamental principles in flowering, circadian, photosynthesis and phytohormone biology (36). The Michael group generated the first high quality genome of a The Greater Duckweed Spirodela polyrhiza, which revealed that Duckweed provide advantages for basic plant biology with a reduced non-redundant set of protein coding genes, no strong centromere arrays and a reduced set of ribosomal arrays (8, 37–39). However, the most morphologically reduced, smallest and fastest growing duckweed Wolffia has the potential to become the yeast of reductionist plant biology. The Michael group published high quality genomes for two clones of W. australiana revealing an even more streamlined genome with only 15,000 non-redundant protein coding genes (32). Coupled to the fact that Wolffia readily flowers, makes crosses, is transformable and requires simple growth conditions, it is currently being developed as a synthetic biology chassis in the Michael group.
sequencing a diverse array of unique plant genomes. Therefore, the Michael group has invested in developing Duckweed as a model system since they are some of the smallest, fastest growing and simplest flowering plants known on Earth (36). Before Arabidopsis was the model plant of choice, plants in the Lemnaceae commonly known as Duckweed were important in reductionist plant biology enabling scientists to establish fundamental principles in flowering, circadian, photosynthesis and phytohormone biology (36). The Michael group generated the first high quality genome of a The Greater Duckweed Spirodela polyrhiza, which revealed that Duckweed provide advantages for basic plant biology with a reduced non-redundant set of protein coding genes, no strong centromere arrays and a reduced set of ribosomal arrays (8, 37–39). However, the most morphologically reduced, smallest and fastest growing duckweed Wolffia has the potential to become the yeast of reductionist plant biology. The Michael group published high quality genomes for two clones of W. australiana revealing an even more streamlined genome with only 15,000 non-redundant protein coding genes (32). Coupled to the fact that Wolffia readily flowers, makes crosses, is transformable and requires simple growth conditions, it is currently being developed as a synthetic biology chassis in the Michael group.
References
1. E. Ibarra-Laclette, E. Lyons, G. Hernández-Guzmán, C. A. Pérez-Torres, L. Carretero-Paulet, T.-H. Chang, T. Lan, A. J. Welch, M. J. A. Juárez, J. Simpson, A. Fernández-Cortés, M. Arteaga-Vázquez, E. Góngora-Castillo, G. Acevedo-Hernández, S. C. Schuster, H. Himmelbauer, A. E. Minoche, S. Xu, M. Lynch, A. Oropeza-Aburto, S. A. Cervantes-Pérez, M. de Jesús Ortega-Estrada, J. I. Cervantes-Luevano, T. P. Michael, T. Mockler, D. Bryant, A. Herrera-Estrella, V. A. Albert, L. Herrera-Estrella, Architecture and evolution of a minute plant genome. Nature. 498, 94–98 (2013).
2. R. Ming, S. Hou, Y. Feng, Q. Yu, A. Dionne-Laporte, J. H. Saw, P. Senin, W. Wang, B. V. Ly, K. L. T. Lewis, S. L. Salzberg, L. Feng, M. R. Jones, R. L. Skelton, J. E. Murray, C. Chen, W. Qian, J. Shen, P. Du, M. Eustice, E. Tong, H. Tang, E. Lyons, R. E. Paull, T. P. Michael, K. Wall, D. W. Rice, H. Albert, M.-L. Wang, Y. J. Zhu, M. Schatz, N. Nagarajan, R. A. Acob, P. Guan, A. Blas, C. M. Wai, C. M. Ackerman, Y. Ren, C. Liu, J. Wang, J. Wang, J.-K. Na, E. V. Shakirov, B. Haas, J. Thimmapuram, D. Nelson, X. Wang, J. E. Bowers, A. R. Gschwend, A. L. Delcher, R. Singh, J. Y. Suzuki, S. Tripathi, K. Neupane, H. Wei, B. Irikura, M. Paidi, N. Jiang, W. Zhang, G. Presting, A. Windsor, R. Navajas-Pérez, M. J. Torres, F. A. Feltus, B. Porter, Y. Li, A. M. Burroughs, M.-C. Luo, L. Liu, D. A. Christopher, S. M. Mount, P. H. Moore, T. Sugimura, J. Jiang, M. A. Schuler, V. Friedman, T. Mitchell-Olds, D. E. Shippen, C. W. dePamphilis, J. D. Palmer, M. Freeling, A. H. Paterson, D. Gonsalves, L. Wang, M. Alam, The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature. 452, 991–996 (2008).
3. International Brachypodium Initiative, Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 463, 763–768 (2010).
4. V. Shulaev, D. J. Sargent, R. N. Crowhurst, T. C. Mockler, O. Folkerts, A. L. Delcher, P. Jaiswal, K. Mockaitis, A. Liston, S. P. Mane, P. Burns, T. M. Davis, J. P. Slovin, N. Bassil, R. P. Hellens, C. Evans, T. Harkins, C. Kodira, B. Desany, O. R. Crasta, R. V. Jensen, A. C. Allan, T. P. Michael, J. C. Setubal, J.-M. Celton, D. J. G. Rees, K. P. Williams, S. H. Holt, J. J. Ruiz Rojas, M. Chatterjee, B. Liu, H. Silva, L. Meisel, A. Adato, S. A. Filichkin, M. Troggio, R. Viola, T.-L. Ashman, H. Wang, P. Dharmawardhana, J. Elser, R. Raja, H. D. Priest, D. W. Bryant Jr, S. E. Fox, S. A. Givan, L. J. Wilhelm, S. Naithani, A. Christoffels, D. Y. Salama, J. Carter, E. Lopez Girona, A. Zdepski, W. Wang, R. A. Kerstetter, W. Schwab, S. S. Korban, J. Davik, A. Monfort, B. Denoyes-Rothan, P. Arus, R. Mittler, B. Flinn, A. Aharoni, J. L. Bennetzen, S. L. Salzberg, A. W. Dickerman, R. Velasco, M. Borodovsky, R. E. Veilleux, K. M. Folta, The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116 (2011).
5. J. A. Banks, T. Nishiyama, M. Hasebe, J. L. Bowman, M. Gribskov, C. dePamphilis, V. A. Albert, N. Aono, T. Aoyama, B. A. Ambrose, N. W. Ashton, M. J. Axtell, E. Barker, M. S. Barker, J. L. Bennetzen, N. D. Bonawitz, C. Chapple, C. Cheng, L. G. G. Correa, M. Dacre, J. DeBarry, I. Dreyer, M. Elias, E. M. Engstrom, M. Estelle, L. Feng, C. Finet, S. K. Floyd, W. B. Frommer, T. Fujita, L. Gramzow, M. Gutensohn, J. Harholt, M. Hattori, A. Heyl, T. Hirai, Y. Hiwatashi, M. Ishikawa, M. Iwata, K. G. Karol, B. Koehler, U. Kolukisaoglu, M. Kubo, T. Kurata, S. Lalonde, K. Li, Y. Li, A. Litt, E. Lyons, G. Manning, T. Maruyama, T. P. Michael, K. Mikami, S. Miyazaki, S.-I. Morinaga, T. Murata, B. Mueller-Roeber, D. R. Nelson, M. Obara, Y. Oguri, R. G. Olmstead, N. Onodera, B. L. Petersen, B. Pils, M. Prigge, S. A. Rensing, D. M. Riaño-Pachón, A. W. Roberts, Y. Sato, H. V. Scheller, B. Schulz, C. Schulz, E. V. Shakirov, N. Shibagaki, N. Shinohara, D. E. Shippen, I. Sørensen, R. Sotooka, N. Sugimoto, M. Sugita, N. Sumikawa, M. Tanurdzic, G. Theissen, P. Ulvskov, S. Wakazuki, J.-K. Weng, W. W. G. T. Willats, D. Wipf, P. G. Wolf, L. Yang, A. D. Zimmer, Q. Zhu, T. Mitros, U. Hellsten, D. Loqué, R. Otillar, A. Salamov, J. Schmutz, H. Shapiro, E. Lindquist, S. Lucas, D. Rokhsar, I. V. Grigoriev, The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 332, 960–963 (2011).
6. R. K. Varshney, W. Chen, Y. Li, A. K. Bharti, R. K. Saxena, J. A. Schlueter, M. T. A. Donoghue, S. Azam, G. Fan, A. M. Whaley, A. D. Farmer, J. Sheridan, A. Iwata, R. Tuteja, R. Varma Penmetsa, W. Wu, H. D. Upadhyaya, S.-P. Yang, T. Shah, K. B. Saxena, T. Michael, W. Richard McCombie, B. Yang, G. Zhang, H. Yang, J. Wang, C. Spillane, D. R. Cook, G. D. May, X. Xu, S. A. Jackson, Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotechnology. 30 (2012), pp. 83–89.
7. R. Ming, R. VanBuren, Y. Liu, M. Yang, Y. Han, L.-T. Li, Q. Zhang, M.-J. Kim, M. C. Schatz, M. Campbell, J. Li, J. E. Bowers, H. Tang, E. Lyons, A. A. Ferguson, G. Narzisi, D. R. Nelson, C. E. Blaby-Haas, A. R. Gschwend, Y. Jiao, J. P. Der, F. Zeng, J. Han, X. J. Min, K. A. Hudson, R. Singh, A. K. Grennan, S. J. Karpowicz, J. R. Watling, K. Ito, S. A. Robinson, M. E. Hudson, Q. Yu, T. C. Mockler, A. Carroll, Y. Zheng, R. Sunkar, R. Jia, N. Chen, J. Arro, C. M. Wai, E. Wafula, A. Spence, Y. Han, L. Xu, J. Zhang, R. Peery, M. J. Haus, W. Xiong, J. A. Walsh, J. Wu, M.-L. Wang, Y. J. Zhu, R. E. Paull, A. B. Britt, C. Du, S. R. Downie, M. A. Schuler, T. P. Michael, S. P. Long, D. R. Ort, J. W. Schopf, D. R. Gang, N. Jiang, M. Yandell, C. W. dePamphilis, S. S. Merchant, A. H. Paterson, B. B. Buchanan, S. Li, J. Shen-Miller, Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14, R41 (2013).
8. W. Wang, G. Haberer, H. Gundlach, C. Gläßer, T. Nussbaumer, M. C. Luo, A. Lomsadze, M. Borodovsky, R. A. Kerstetter, J. Shanklin, D. W. Byrant, T. C. Mockler, K. J. Appenroth, J. Grimwood, J. Jenkins, J. Chow, C. Choi, C. Adam, X.-H. Cao, J. Fuchs, I. Schubert, D. Rokhsar, J. Schmutz, T. P. Michael, K. F. X. Mayer, J. Messing, The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 5, 3311 (2014).
9. R. VanBuren, D. Bryant, P. P. Edger, H. Tang, D. Burgess, D. Challabathula, K. Spittle, R. Hall, J. Gu, E. Lyons, M. Freeling, D. Bartels, B. Ten Hallers, A. Hastie, T. P. Michael, T. C. Mockler, Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 527, 508–511 (2015).
10. R. VanBuren, D. Bryant, J. M. Bushakra, K. J. Vining, P. P. Edger, E. R. Rowley, H. D. Priest, T. P. Michael, E. Lyons, S. A. Filichkin, M. Dossett, C. E. Finn, N. V. Bassil, T. C. Mockler, The genome of black raspberry (Rubus occidentalis). Plant J. 87, 535–547 (2016).
11. 1001 Genomes Consortium. Electronic address: magnus.nordborg@gmi.oeaw.ac.at, 1001 Genomes Consortium, 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell. 166, 481–491 (2016).
12. R. VanBuren, C. M. Wai, S. Ou, J. Pardo, D. Bryant, N. Jiang, T. C. Mockler, P. Edger, T. P. Michael, Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla. Nat. Commun. 9, 13 (2018).
13. T. P. Michael, F. Jupe, F. Bemm, S. T. Motley, J. P. Sandoval, C. Lanz, O. Loudet, D. Weigel, J. R. Ecker, High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 9, 541 (2018).
14. P. N. T. Hoang, T. P. Michael, S. Gilbert, P. Chu, S. T. Motley, K. J. Appenroth, I. Schubert, E. Lam, Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J. 96, 670–684 (2018).
15. F. Jupe, A. C. Rivkin, T. P. Michael, M. Zander, S. T. Motley, J. P. Sandoval, R. K. Slotkin, H. Chen, R. Castanon, J. R. Nery, J. R. Ecker, The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 15, e1007819 (2019).
16. J. R. Chekan, S. M. K. McKinnie, M. L. Moore, S. G. Poplawski, T. P. Michael, B. S. Moore, Scalable Biosynthesis of the Seaweed Neurochemical, Kainic Acid. Angew. Chem. Int. Ed Engl. 58, 8454–8457 (2019).
17. C. M. Wai, S. E. Weise, P. Ozersky, T. C. Mockler, T. P. Michael, R. VanBuren, Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album. PLoS Genet. 15, e1008209 (2019).
18. T. Kawakatsu, S.-S. C. Huang, F. Jupe, E. Sasaki, R. J. Schmitz, M. A. Urich, R. Castanon, J. R. Nery, C. Barragan, Y. He, H. Chen, M. Dubin, C.-R. Lee, C. Wang, F. Bemm, C. Becker, R. O’Neil, R. C. O’Malley, D. X. Quarless, 1001 Genomes Consortium, N. J. Schork, D. Weigel, M. Nordborg, J. R. Ecker, Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell. 166, 492–505 (2016).
19. C. J. Grassa, G. D. Weiblen, J. P. Wenger, C. Dabney, S. G. Poplawski, S. Timothy Motley, T. P. Michael, C. J. Schwartz, A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 230, 1665–1679 (2021).
20. M. Naish, M. Alonge, P. Wlodzimierz, A. J. Tock, B. W. Abramson, A. Schmücker, T. Mandáková, B. Jamge, C. Lambing, P. Kuo, N. Yelina, N. Hartwick, K. Colt, L. M. Smith, J. Ton, T. Kakutani, R. A. Martienssen, K. Schneeberger, M. A. Lysak, F. Berger, A. Bousios, T. P. Michael, M. C. Schatz, I. R. Henderson, The genetic and epigenetic landscape of the Arabidopsis centromeres. Science. 374, eabi7489 (2021).
21. R. VanBuren, C. Man Wai, X. Wang, J. Pardo, A. E. Yocca, H. Wang, S. R. Chaluvadi, G. Han, D. Bryant, P. P. Edger, J. Messing, M. E. Sorrells, T. C. Mockler, J. L. Bennetzen, T. P. Michael, Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat. Commun. 11, 884 (2020).
22. D. Wickell, L.-Y. Kuo, H.-P. Yang, A. D. Ashok, I. Irisarri, A. Dadras, S. de Vries, J. de Vries, Y.-M. Huang, Z. Li, M. S. Barker, N. T. Hartwick, T. P. Michael, F.-W. Li, Underwater CAM photosynthesis elucidated by Isoetes genome. bioRxiv (2021), p. 2021.06.09.447806.
23. B. N. Mansfeld, A. Boyher, J. C. Berry, M. Wilson, S. Ou, S. Polydore, T. P. Michael, N. Fahlgren, R. S. Bart, Large structural variations in the haplotype-resolved African cassava genome. Plant J. (2021), doi:10.1111/tpj.15543.
24. T. P. Michael, P. A. Salomé, H. J. Yu, T. R. Spencer, E. L. Sharp, M. A. McPeek, J. M. Alonso, J. R. Ecker, C. R. McClung, Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 302, 1049–1053 (2003).
25. T. P. Michael, C. R. McClung, Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 130, 627–638 (2002).
26. T. P. Michael, C. R. McClung, Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132, 629–639 (2003).
27. T. P. Michael, P. A. Salome, C. R. McClung, Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci. U. S. A. 100, 6878–6883 (2003).
28. T. P. Michael, G. Breton, S. P. Hazen, H. Priest, T. C. Mockler, S. A. Kay, J. Chory, A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6, e225 (2008).
29. T. C. Mockler, T. P. Michael, H. D. Priest, R. Shen, C. M. Sullivan, S. A. Givan, C. McEntee, S. A. Kay, J. Chory, The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72, 353–363 (2007).
30. T. P. Michael, T. C. Mockler, G. Breton, C. McEntee, A. Byer, J. D. Trout, S. P. Hazen, R. Shen, H. D. Priest, C. M. Sullivan, S. A. Givan, M. Yanovsky, F. Hong, S. A. Kay, J. Chory, Network Discovery Pipeline Elucidates Conserved Time-of-Day–Specific cis-Regulatory Modules. PLoS Genet. 4, e14 (2008).
31. S. A. Filichkin, G. Breton, H. D. Priest, P. Dharmawardhana, P. Jaiswal, S. E. Fox, T. P. Michael, J. Chory, S. A. Kay, T. C. Mockler, Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One. 6, e16907 (2011).
32. T. P. Michael, E. Ernst, N. Hartwick, P. Chu, D. Bryant, S. Gilbert, S. Ortleb, E. L. Baggs, K. S. Sree, K. J. Appenroth, J. Fuchs, F. Jupe, J. P. Sandoval, K. V. Krasileva, L. Borisjuk, T. C. Mockler, J. R. Ecker, R. A. Martienssen, E. Lam, Genome and time-of-day transcriptome of Wolffia australiana link morphological minimization with gene loss and less growth control. Genome Res. (2020), doi:10.1101/gr.266429.120.
33. A. Zdepski, W. Wang, H. D. Priest, F. Ali, M. Alam, T. C. Mockler, T. P. Michael, Conserved Daily Transcriptional Programs in Carica papaya. Trop. Plant Biol. 1, 236–245 (2008).
34. K. J.-M. MacKinnon, B. J. Cole, C. Yu, J. H. Coomey, N. T. Hartwick, M.-S. Remigereau, T. Duffy, T. P. Michael, S. A. Kay, S. P. Hazen, Changes in ambient temperature are the prevailing cue in determining Brachypodium distachyon diurnal gene regulation. New Phytol. (2020), doi:10.1111/nph.16507.
35. T. P. Michael, Core circadian clock and light signaling genes brought into genetic linkage across the green lineage, , doi:10.1101/2021.11.02.466975.
36. K. Acosta, K. J. Appenroth, L. Borisjuk, M. Edelman, U. Heinig, M. A. K. Jansen, T. Oyama, B. Pasaribu, I. Schubert, S. Sorrels, K. S. Sree, S. Xu, T. P. Michael, E. Lam, Return of the Lemnaceae: duckweed as a model plant system in the genomics and postgenomics era. Plant Cell. 33, 3207–3234 (2021).
37. P. T. N. Hoang, A. Fiebig, P. Novák, J. Macas, H. X. Cao, A. Stepanenko, G. Chen, N. Borisjuk, U. Scholz, I. Schubert, Chromosome-scale genome assembly for the duckweed Spirodela intermedia, integrating cytogenetic maps, PacBio and Oxford Nanopore libraries. Sci. Rep. 10, 19230 (2020).
38. T. P. Michael, D. Bryant, R. Gutierrez, N. Borisjuk, P. Chu, H. Zhang, J. Xia, J. Zhou, H. Peng, M. El Baidouri, B. Ten Hallers, A. R. Hastie, T. Liang, K. Acosta, S. Gilbert, C. McEntee, S. A. Jackson, T. C. Mockler, W. Zhang, E. Lam, Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89, 617–635 (2017).
39. A. Harkess, F. McLoughlin, N. Bilkey, K. Elliott, R. Emenecker, E. Mattoon, K. Miller, K. Czymmek, R. D. Vierstra, B. C. Meyers, Others, Improved Spirodela polyrhiza genome and proteomic analyses reveal a conserved chromosomal structure with high abundance of chloroplastic proteins favoring energy production. J. Exp. Bot. 72, 2491–2500 (2021).